Introduction to Linux
These lessons will focus on drawing together and supplementing existing resources on using Bash and common command line utilities. The Bash shell is typically used as part of the Linux or MacOS operating systems and while both of these operating systems share a common origin in the Unix of the 1970s, 80s and 90s, there are also some differences between Linux and MacOS that we will note as we go along.
Before we start - go to Mentimeter to join the presentation.
Tasks
We will be focusing on tasks using the resources listed in the Resources section. To get started open the Terminal application so that you can be in a bash shell environment.
How to set up your environment if you are not part of the course at SANBI
This lesson was written for an in-person course at SANBI but, except for the “Working with Remote Computers” part of the lesson, the practicals can be conducted on any computer with a typical Linux or MacOS X environment. The practicals assume that you have a folder called data in your home directory with a few small files in it. To create this folder with the files, run this command:
curl https://pathogen-genomics-march-2024.sanbi.ac.za/data/shell/make_lesson_data.sh |bash
This command uses curl to download a script and then runs the script using bash. In general be very wary of instructions that require you to run an unknown script on your computer, but in this case, if you trust the authors of this training material, you can trust this script.
The above command should do a few things and then print DONE to the screen. If you have seen that DONE you can proceed to Task 1.
Task 1: Exploring using Linux commands
In your web browser, go to the shell server. You will have been given a username and password to log in with.
First, orient yourself. How do you find out where you are?
Show answer
pwdLook around. What can you see? How can you tell if items are files or directories?
Show answer
The ls command has two useful flags that can distinguish files and directories. For a
long listing:
ls -l
and for a short listing but with classification indicators use ls -F
Now that you know that there is a directory called data/ in your home directory, how
can you see what is in it?
Show answer
You can list the directory contents by naming it, e.g.
ls data/
or by changing your current directory to it and running ls, i.e. cd data followed
by ls. In either case you can see that there is a file called cases.csv in this
directory.
Next, how can you see what is in a file? There are a number of different ways, some a
little safer than others. You might recall that cat can read a file. What are some
disadvantages of peeking into a file using cat? And what are some alternatives?
Show answer
The cat command reads the whole file, which might not be what you want if you are
dealing with a large file. If you see page upon page of data scrolling past your
screen, you can stop the cat command (or, in general, any other command) by hitting
the key combination Ctrl-C.
A second downside to using cat to look into files is that if you happen to use it on
a binary file you see a whole lot of strange characters.
One alternative to cat is to use head. This won’t scroll off your screen but
will still display junk if you display a binary file.
Finally you can use the more command. The more command (and, if it is available,
the very similar less command) let you page through a file and will warn you if
you are trying to view a binary file.
case_id,department,case_date,gender,age,age_unit
CAS00001,Centre,2015-01-16,F,5,years
CAS00002,Centre,2015-02-23,M,7,months
CAS00003,Nord-Est,2015-01-07,F,12,years
CAS00004,Ouest,2015-01-18,F,39,years
The cases.csv file contains some simulated data on cholera cases in Haiti. Once eradicated
from the island, cholera reappeared in the Department (or region) of Artibonite in 2010 and
more than 800,000 cases of disease have been recorded over the past decade.
If you’ve got this far, you’ve found that you’ve been provided with a file on cholera case data, from the island state of Haiti.
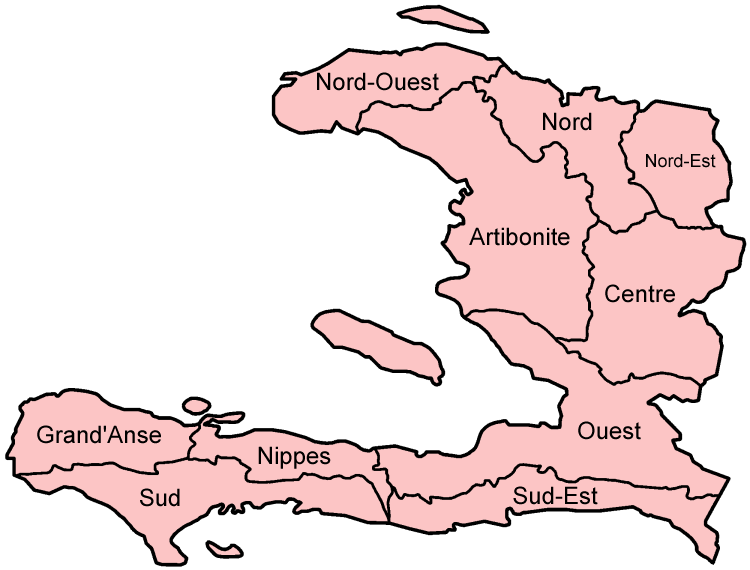
How many cases are recorded in the file? What are the headings? In the case ID, why was the location where the case was collected not encoded as part of the ID?
Now that we’ve got some idea of the structure of the file, let’s use shell commands to answer a few more questions. First, how many unique locations (departments) are listed in the file?
To answer this question, you need to extract the departments column (the 2nd column) and find out how many unique values are in that column.
There are two common ways to extract a column from a file, one using cut and one using awk. In both cases these tools are designed with pipes in mind, so
they can either read from a file or from stdin. And in both cases you need to take note of what the field separator is: in this case it is a comma (“,”),
making this file a Comma Separated Values (CSV). Another common file type that you might encounter is a TSV, where columns are separated with tabs.
Of the two tools, cut is the simpler, and it uses -d to set the delimiter (which is a tab by default) and -f to select a field:
cut -d, -f2 cases.tsv
The awk tools is more powerful but also more complex. It is very well worth learning how to use awk and there are good guides online (including some of those
listed as part of our course resources). For awk we also need to specific a field separator, with -F, and then a small program to extract the field we want.
The program { print $2} prints the 2nd field for each line in the file. Notice that it uses variables with $ signs, just like the shell, so to stop
the shell interpreting our awk program we have to specific it in single quotes:
awk -F, '{print $2}'
If you look closely, you’ll notice that both cut and awk shows us the first line of the file (the header) and we don’t want to see that. While we could use some
awk programming to avoid that, the simplest way to start reading from the second line onwards is using tail with the -n+2 flag. If you specify locations with
a +, tail will start reading from that line onwards, instead of counting backwards from the end of the file. A pipe lets us combine the two commands:
tail -n+2 cases.csv |cut -d, -f2
Show answer
The first part of the answer shows us how to extract the data we need from the file. Now, how to to count the unique values?
In the shell, sort and uniq let us turn a list of values into a list of distinct values. And wc lets us count, so these three programs form part
of the everyday vocabulary of shell users.
The uniq command only works on repeated lines, i.e. lines that are right next to each other. This is why we need to ensure that data is sorted with sort
before using uniq.
Show answer
Putting it all together:
tail -n+2 cases.csv |cut -d, -f2 |sort |uniq
will show us the unique values in column 2 and:
tail -n+2 cases.csv |cut -d, -f2 |sort |uniq|wc -l
will count how many unique departments there are in our dataset (there are 9).
The third column of the dataset is the date that the case was collected. This is organised in YEAR-MONTH-DAY order with 0-padded months and days, which aligns with the ISO 8601 standard. One advantage of this way of writing dates is that they are easily sorted.
Using this information, how can we find the maximum date recorded in the file?
Show answer
The tool to use here is sort. On its own sort can focus on a particular column (known as a key). Once again we need to tell the tool how to separate
columns, this time using the -t flag. So as a first attempt:
sort -t, -k3 cases.csv
works to sort the data by date, from earliest to latest. Again, we probably want to use tail -n+2 to skip the header.
There is another thing to note about -k3: the default sort order used by sort is dictionary sort. For this data, that doesn’t make a difference, but
if we want to sort numbers, we need to use -k3n. If our dates were expressed without the leading zeros it would cause a problem for sort. Luckily our
ISO-8601-compliant dates don’t have this problem.
Again we might want to combine a few commands:
tail -n+2 cases.csv |cut -d, -f3 |sort |tail -1
What answer does that give us?
To find the minimum date, we need a few small changes? What are they
Show answer
We could either reverse the sort order with -r:
tail -n+2 cases.csv |cut -d, -f3 |sort -r |tail -1
or we could pick the first line from the original smallest-to-largest sort:
tail -n+2 cases.csv |cut -d, -f3 |sort |head -1
Shifting focus again, how can we find out which department has the most cases?
Show answer
The uniq tool has a -c flag for counting how often distinct values appear. We can use that here:
tail -n+2 cases.csv |cut -d, -f2 | sort | uniq -c
Remember, always sort before uniq!
Male and female gender is encoded as M or F in the dataset. Thank goodness that this is clean data! Which gender saw the most cases?
Show answer
We can use a very similar set of commands to answer this question, now just using column 4:
tail -n+2 cases.csv |cut -d, -f4 | sort | uniq -c
Finally, how many cases were seen in children under 1 year of age? We have to look carefully at the data to answer this.
Show answer
The age is expressed in months for children under age 1. We can use that to filter out the data we need using the grep command:
tail -n+2 cases.csv |cut -d, -f6 | grep months | wc -l
Or we could use awk. Once again awk is a bit more complex but more flexible:
tail -n+2 cases.csv |awk -F, '$6 ~ /months/ {tot = tot + 1} END {print tot}'
The test used in awk here is actually a regular expression (i.e. pattern) match so we can use it to check for more than just the word “months”.
Resources
Terminal Basics from Sandbox.Bio
Bashcrawl - an adventure in Bash exploration